The R Function of the Day series will focus on describing in plain language how certain R functions work, focusing on simple examples
that you can apply to gain insight into your own data.
Today, I will discuss the foodweb function, found in the mvbutils
package.
Foodweb? What is that?
In biology, a foodweb is a group of food chains, showing the complex
relationships that exist in nature. Similarly, the R foodweb
function contained in the mvbutils package on CRAN displays a
flowchart of function calls. What functions does my function call?
What funtions in my package call the lm function? These are the
types of questions that can be answered in a graphical way using
foodweb. This information can be useful for documenting your own
code, and for learning how a package that you’re not familiar with
works. At the end of this post, you’ll see an example with a diagram
of the survexp function from the survival package.
First, you have to install and load the mvbutils package, so let’s
do that first.
install.packages("mvbutils") library(mvbutils)
A simple example
Let’s define a couple simple functions to see how foodweb works. We
simply define a function called outer that calls two functions,
inner1 and inner2.
inner1 <- function() { "This is the inner1 function" } inner2 <- function() { "This is the inner2 function" } outer <- function(x) { i1 <- inner1() i2 <- inner2() }
Now let’s use the foodweb function to diagram the relationship between
the outer and inner functions. If we don’t give foodweb any
arguments, it will scour our global workspace for functions, and make
the diagram. Since the only functions in my global workspace are the
ones we’ve just defined, we get the following plot.
foodweb()
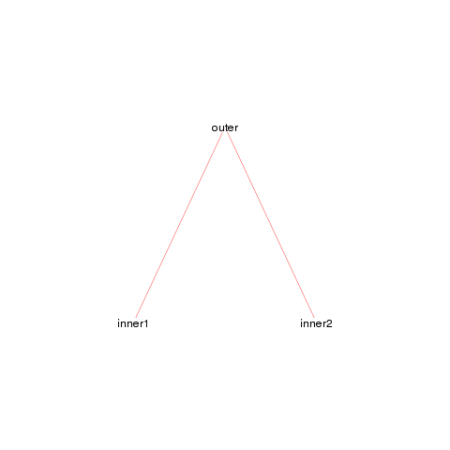
We see a simple graph showing that the outer function calls both the
inner1 and inner2 functions, jus as we expect. We can make this look a
bit nicer by adjusting a few of the arguments.
foodweb(border = TRUE, expand.xbox = 3, boxcolor = "#FC6512", textcolor = "black", cex = 1.2, lwd=2)
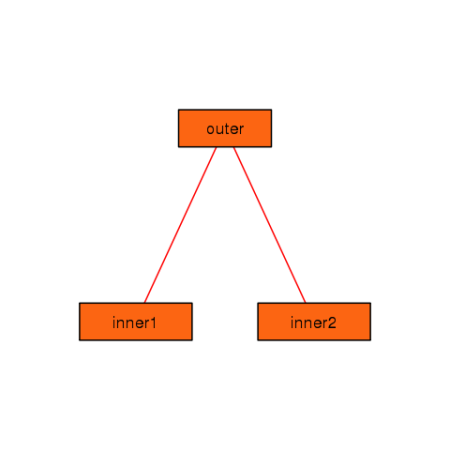
You can see that you can control many of the graphical parameters of
the resulting plot. See the help page for foodweb to see the
complete list of graphical parameters you can specify.
A more complicated example
As we saw above, by default foodweb will look in your global
workspace for functions to construct the web from. However, you can
pass foodweb a group of functions to operate on. There are several
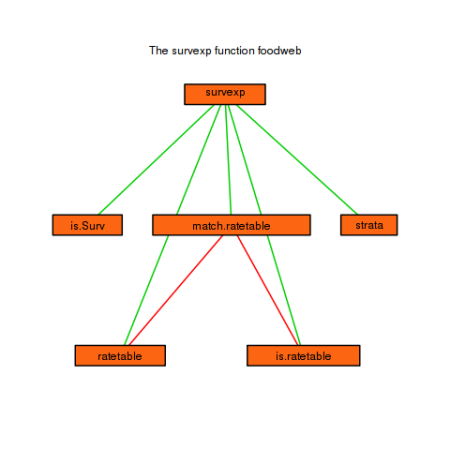
ways of doing this, see the help page for examples. The code below
shows one possiblity, when we want to limit our results to functions
appearing in a specific package. The prune argument is very useful.
It takes a string (or regular expression) to prune the resulting graph
to.
foodweb(where = "package:survival", prune = "survexp", border = TRUE, expand.xbox = 2, boxcolor = "#FC6512", textcolor = "black", cex = 1.0, lwd=2) mtext("The survexp function foodweb")
Conclusion
I have used the foodweb function to help understand other user’s
code that I have inherited. It has proved very valuable in aiding the
comprehension of hard to read or complicated functions. I have also
used it in the development of my own packages, as it sometimes can
suggest ways to reorganize the code to be more logical.
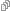

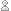 Posted by erikr
Posted by erikr